-
PyTorch Example of LSTMData/Machine learning 2021. 4. 21. 15:19
Architecture [3]
The main components are: 1) hidden and cell states, 2) input gate, forget gate, output gate.
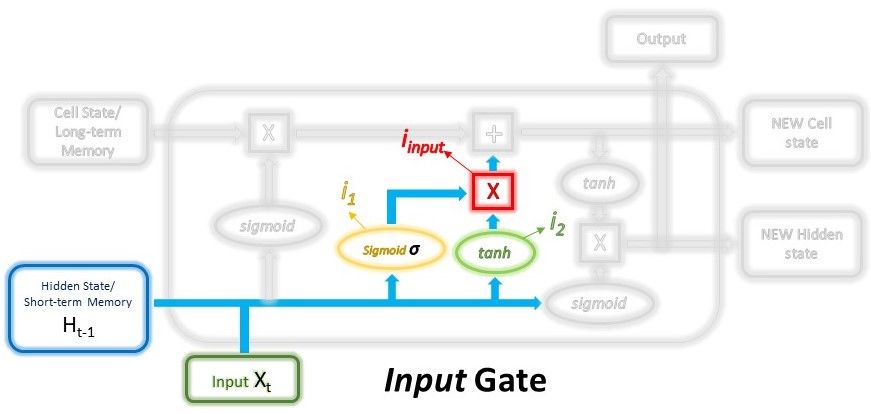
$$ i_1 = \sigma ( W_{i_1} \cdot (H_{t-1}, x_t) + b_{i_1} ) $$
$$ i_ = tanh ( W_{i_2} \cdot (H_{t-1}, x_t) + b_{i_2} ) $$
$$ i_{input} = i_1 * i_2 $$
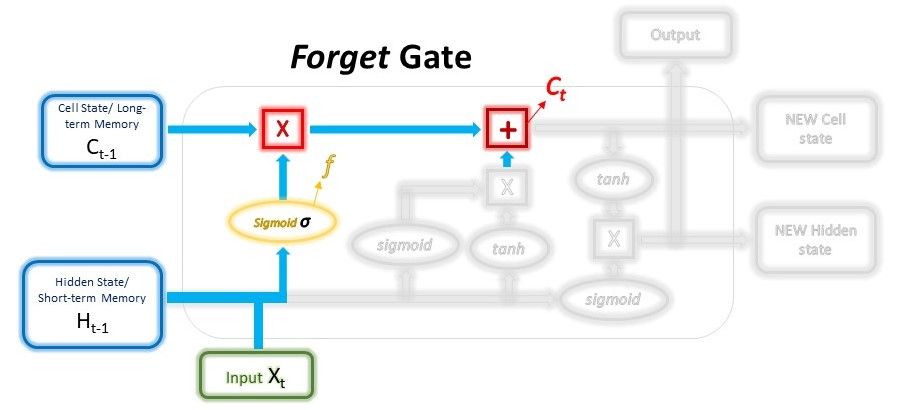
$$ f = \sigma ( W_{forget} \cdot (H_{t-1}, x_t) + b_{forget} ) $$
$$ C_t = C_{t-1} * f + i_{input} $$
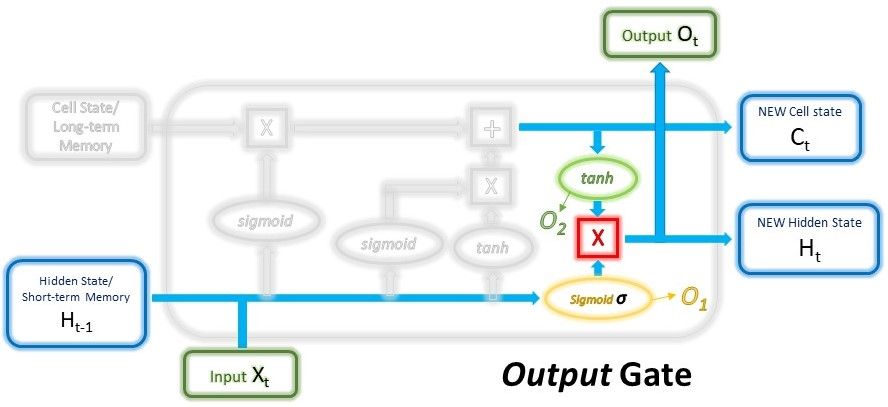
$$ O_1 = \sigma ( W_{output_1} \cdot (H_{t-1}, x_t) + b_{output_1} ) $$
$$ O_2 = tanh ( W_{output_2} \cdot C_t + b_{output_2} ) $$
$$ H_t, O_t = O_1 * O_2 \tag{1} $$
In $Eq. (1)$, $H_t == Q_t$ is valid only when there is one LSTM layer.
PyTorch Example [1, 2, 4]
An LSTM model is trained to predict a gradient of first-order linear data. For example, if data is from $y=0.3 x$, the model should predict $0.3$.
Dataset
A (mini-)batch's dimension needs to be specially organized for LSTM as $(batch\_size, seq\_len, input\_size)$ if
batch_first=Trueinnn.LSTM(..). Data from each linear function is organized as follows: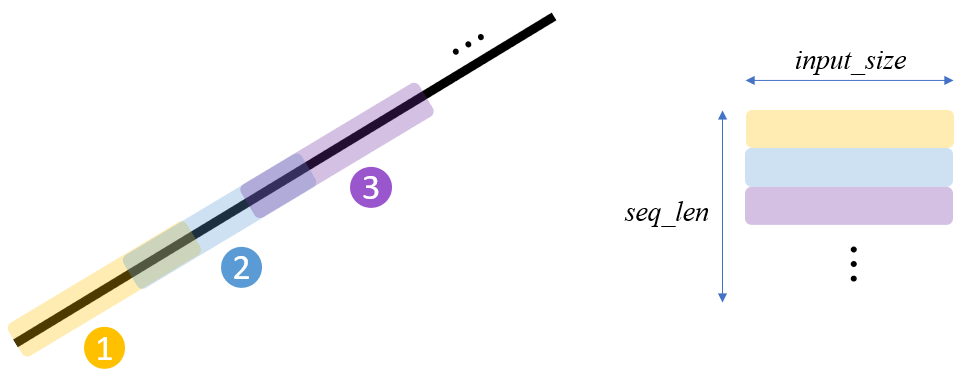
By fetching this stacked data over the batch dimension via DataLoder, we can obtain a training dataset for LSTM. Note that different linear gradients are used to stack along the batch dimension.
import numpy as np import matplotlib.pyplot as plt import torch import torch.nn as nn from torch.utils.data import Dataset, DataLoader # generate a dataset class MyDataset(Dataset): def __init__(self, n_grads, H): super().__init__() self.grads = np.random.uniform(-1, 1, size=n_grads) self.x = torch.arange(0, 1, 0.01) self.H = H # horizon [steps] self.interval = H//2 # [steps] self.len = n_grads def __getitem__(self, idx): """ 1. sample `grads` 2. generate linear data (seq_len*input_size) Remarks: (batch_size*seq_len*input_size) is formed by DataLoader. """ H = self.H # 1. grad = self.grads[idx] # 2. y = grad * self.x subys = torch.tensor([]) # (seq_len*input_size(H)) i = 0 while len(y[i:i+H]) == H: suby = y[i:i+H].view(1, -1) subys = torch.cat((subys, suby), 0) i += self.interval return subys, grad def __len__(self): return self.lenModel
The LSTM layer receives the mini-batch ($(batch\_size, seq\_len, input\_size)$) and yieds an output ($(batch\_size, seq\_len, hidden\_size)$). Then, we choose the last $output$ w.r.t the $seq\_len$ dimension, and it is fed into a linear layer for the regression task.
One important thing is to initialize the states (hidden and cell states) as zeros in the beginning w.r.t the $seq\_len$ dimension during training.
class LSTM(nn.Module): def __init__(self, in_size): super().__init__() self.in_size = in_size self.h_size = 32 self.n_layers = 2 # define the LSTM layer self.lstm = nn.LSTM(self.in_size, self.h_size, self.n_layers, batch_first=True) self.h_n = None # hidden state self.c_n = None # cell state # define the output layer self.linear = nn.Linear(self.h_size, 1) # predicts `linear-grad` def init_states(self, batch_size): """initialize the hidden and cell states""" self.h_n = torch.zeros(self.n_layers, batch_size, self.h_size) self.c_n = torch.zeros(self.n_layers, batch_size, self.h_size) def forward(self, x): """ x: (batch, seq_len, input_size) out: (batch, seq_len, hidden_size) h_n, c_n: (num_layers * num_directions, batch, hidden_size) """ states = (self.h_n, self.c_n) out, (self.h_n, self.c_n) = self.lstm(x, states) # get the last one of the (sequential) output from the LSTM out = out[:, -1, :] # (batch, hidden_size) out = self.linear(out) # (batch, 1) return outDataLoader & Model & Compile
# DataLoader train_dataset = MyDataset(n_grads=30, H=10) train_data_loader = DataLoader(train_dataset, batch_size=8) val_dataset = MyDataset(n_grads=10, H=10) val_data_loader = DataLoader(val_dataset, batch_size=8) # Model model = LSTM(in_size=train_dataset.H) # Compile optimizer = torch.optim.Adam(model.parameters(), lr=1e-3, weight_decay=1e-5) criterion = nn.MSELoss()Train
Note that .eval() or .train() are not used since there is no layer that requires that. Also, note that
model.init_states(..)is used at the beginning of each mini-batch.# Train n_epochs = 100 for epoch in range(1, n_epochs+1): # train i, train_loss, val_loss = 0, 0., 0. for x, y in train_data_loader: optimizer.zero_grad() model.init_states(batch_size=x.shape[0]) out = model(x) loss = criterion(out, y.float().view(-1, 1)) train_loss += loss.item() loss.backward() optimizer.step() i += 1 train_loss /= i # validate with torch.no_grad(): i = 0 for x, y in val_data_loader: model.init_states(batch_size=x.shape[0]) out = model(x) loss = criterion(out, y.float().view(-1, 1)) val_loss += loss.item() i += 1 val_loss /= i print('epoch: {} | train_loss: {:0.5f} | val_loss: {:0.5f}'.format(epoch, train_loss, val_loss))Results
Training
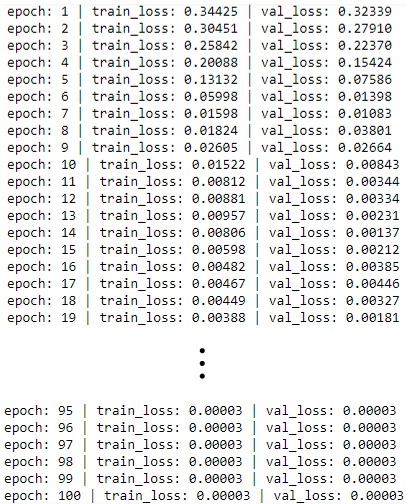
Test
# Test with torch.no_grad(): for x, y in val_data_loader: model.init_states(batch_size=x.shape[0]) out = model(x) break #loss = criterion(out, y.float().view(-1, 1)) # Plot plt.plot(y.float(), 'o', label='y') plt.plot(out.view(-1), '^', label='yhat') plt.legend();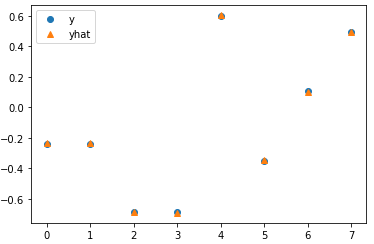
References
[1] Yung, J., 2018, "LSTMs for Time Series in PyTorch" (link)
[2] PyTorch LSTM example (link)
[3] Loye G., 2019, "Long Short-Term Memory: From Zero to Hero with PyTorch" (link)
[4] PyTorch: LSTM (link)'Data > Machine learning' 카테고리의 다른 글
Metrics for Multi-label classification (0) 2021.05.03 Log-bilinear Language Model (0) 2021.04.20 Visualization of CNN (0) 2021.04.15