-
Gower's DistanceData/Machine learning 2021. 1. 8. 17:16
It measures the dissimilarity between records within a mixed dataset. The mixed dataset refers to a dataset where both a numeric feature (variable) and a categorical feature (variable) exist together.
The basic idea behind Gower's distance is to apply a different distance metric to each variable depending on the type of data:
- For numeric variables and ordered factors (ordered categorical variable), distance is calculated as the absolute value of the difference between two records (Manhattan distance).
- For categorical variables, the distance is 1 if the categories between two records are different and the distance is 0 if the categories are the same (Dice distance).
The mathematical formula for the Gower's distance is:
$$ S_{ij} = \frac{\sum_k{w_{ijk} S_{ijk}}}{ \sum_k w_{ijk} } $$
where $S_{ij}$ is the dissimilarity matrix w.r.t records, $S_{ijk}$ is the dissimilarity matrix of a $k$-th variable w.r.t records, $w_{ijk}$ is weight for each dissimilarity matrix $S_{ijk}$.
Algorithm by Example
Let's say we have the following example dataset:
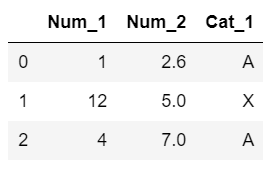
Dataset Since there are three records, our dissimilarity matrix $S_{ij}$ and $S_{ijk}$ will be $3 \times 3$ matrices. Here are the simple steps for the algorithm:
- Compute three $S_{ijk}$ where $k=1, 2, 3$ since there are three variables.
- Add $S_{ij1}, S_{ij2}, S_{ij3}$ together, either using a simple mean or weighted mean.
Note that the Manhattan distance is used for the numeric variables and the dice distance is used for the categorical variable.
First, we compute $S_{ij1}$ by
sklearn.neighbors.DistanceMetric.get_metric("manhattan").pairwise(df[['Num_1']]), and normalize the dissimilarity matrix to the range 0 to 1:
Dissimilarity matrix $S_{ij1}$ 
Normalized dissimilarity matrix Second, we compute $S_{ij2}$ by
DistanceMEtric.get_metric("manhattan"), and normalize it to the range 0 to 1:
Normalized dissimilarity matrix Third, we compute $S_{ij3}$. In this case, we have to compute the dice distance, since the third variable is a categorical variable. The dice distance is 0 whenever the values are equal, and 1 otherwise. The detailed computation of the dice distance can be found here. With
DistanceMetric.get_metric("dice").pairwise(\alpha)where $\alpha$ refers to the data corresponding to 'Cat_1' that's converted into a numeric variable. Then, you get:
Dice distance of the categorical variable. Note that the categorical variable must've been converted into a numeric variable to run the dice distance The final step is to add $S_{ij1}, S_{ij2}, S_{ij3}$ together, either using a simple mean or weighted mean. If we use the simple mean, the result is:

Dissimilarity matrix by the Gower's distance The interpretation of the resulting dissimilarity matrix is:
- The distance between Row1 and Row2 is 0.84 and that of between Row1 and Row3 is 0.42.
- This means Row1 is more similar to Row3 compared to Row2. Intuitively, this makes sense as if we take a look at the dataset.
Application
You can apply hierarchical clustering (e.g. Agglomerative algorithm) to the resulting dissimilarity matrix. Refer to the steps of the agglomeration algorithm:
- Create an initial set of clusters with each cluster consisting of a single record for all records in the data.
- Compute the dissimilarity between all pairs of clusters.
- Merge the two clusters that are least dissimilar as measured by the dissimilarity metric. $\rightarrow$ This is where we can utilize the dissimilarity matrix we computed by the Gower's distance.
- Repeat steps 1-3 until we are left with one remaining cluster.
One Line Implementation
There is a python library, gower, to compute the Gower's distance. The implementation is one simple line:
gower.gower_matrix(df)Source: here
'Data > Machine learning' 카테고리의 다른 글
Problems with Clustering Mixed Data (0) 2021.01.10 Outlier Detection with Multivariate Normal Distribution (0) 2021.01.08 Hierarchical Clustering (Agglomerative Algorithm) (0) 2021.01.07